VELOCIraptor Integration
swiftsimio can be used with the velociraptor library
to extract the particles contained within a given halo and its surrounding
region.
The velociraptor library has documentation also available on
ReadTheDocs here. It can
be installed from PyPI using pip install velociraptor.
The overarching workflow for this integration is as follows:
Load the halo catalogue and groups file using the
velociraptormodule.Get two objects, corresponding to the bound and unbound particles, for a halo.
Use the to_swiftsimio_dataset to load the region around the halo with our ahead-of-time masking technique.
Use the region around the halo directly, or use the mask provided for each particle type to only consider bound particles.
This workflow is explored below. You can use the example data available below if you do not have any SWIFT and VELOCIraptor data available.
http://virgodb.cosma.dur.ac.uk/swift-webstorage/IOExamples/small_cosmo_volume.zip
Example
First, we must load the VELOCIraptor catalogue as follows:
from velociraptor import load as load_catalogue
from velociraptor.particles import load_groups
catalogue_name = "velociraptor"
snapshot_name = "snapshot"
catalogue = load_catalogue(f"{catalogue_name}.properties")
groups = load_groups(f"{catalogue_name}.catalog_groups", catalogue=catalogue)
Then, to extract the largest halo in the volume
particles, unbound_particles = groups.extract_halo(halo_id=0)
To load the particles to a swiftsimio dataset,
from velociraptor.swift.swift import to_swiftsimio_dataset
data, mask = to_swiftsimio_dataset(
particles,
f"{snapshot_name}.hdf5",
generate_extra_mask=True
)
with the generate_extra_mask providing the second return value which
is a mask to extract only the bound particles in the system.
Making an image of the full box shows that only a small subsection of the volume has been loaded (those within twice the maximal usable radius within VELOCIraptor)
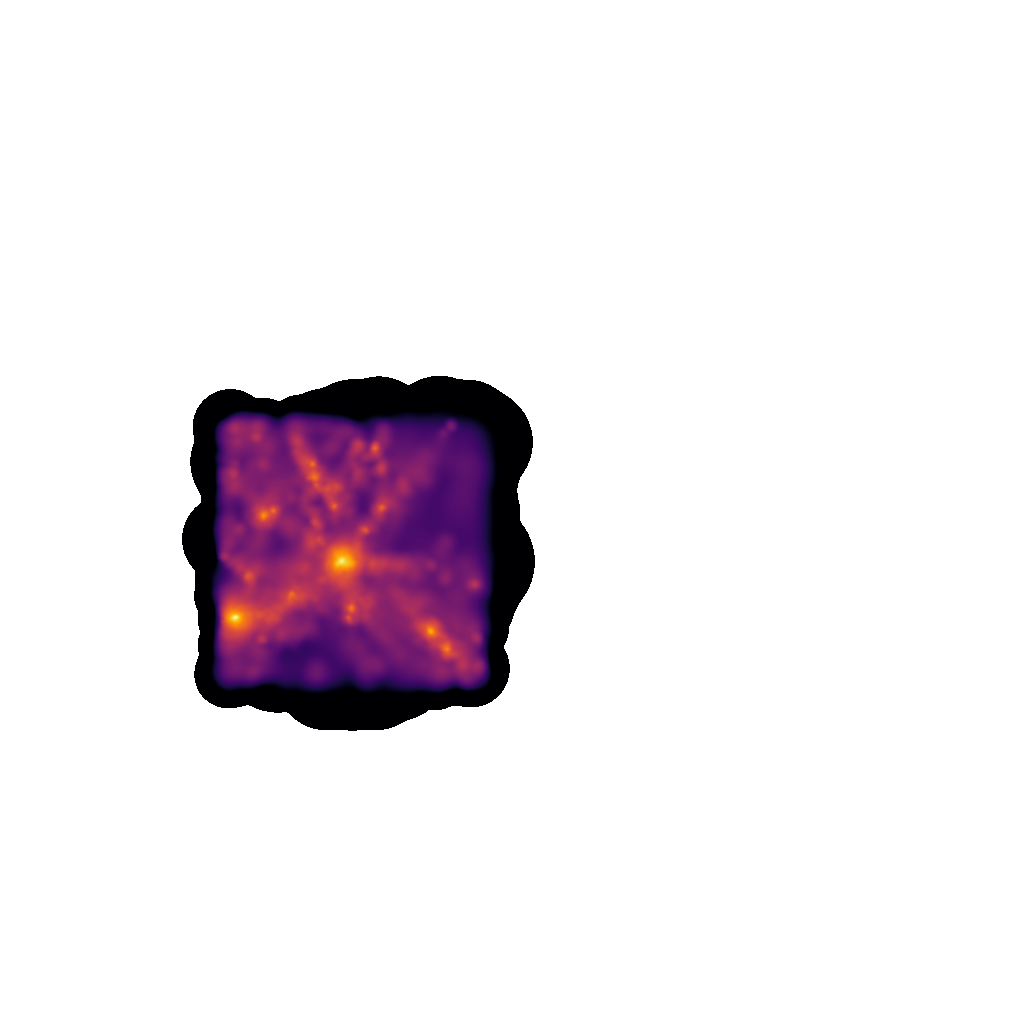
The code for making this image is as follows:
from swiftsimio.visualisation import project_gas_pixel_grid
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
grid = project_gas_pixel_grid(data=data, resolution=1024)
fig, ax = plt.subplots(figsize=(4, 4), dpi=1024 // 4)
fig.subplots_adjust(0, 0, 1, 1)
ax.axis("off")
ax.imshow(grid.T, origin="lower", cmap="inferno", norm=LogNorm(vmin=1e4, clip=True))
fig.savefig("load_halo_fullbox.png")
To make an image of just the central halo, we can access properties on the
particles instance to get the position of the halo.
region = [
particles.x_mbp - particles.r_200crit, particles.x_mbp + particles.r_200crit,
particles.y_mbp - particles.r_200crit, particles.y_mbp + particles.r_200crit,
]
grid = project_gas_pixel_grid(data=data, resolution=1024, region=region)
fig, ax = plt.subplots(figsize=(4, 4), dpi=1024 // 4)
fig.subplots_adjust(0, 0, 1, 1)
ax.axis("off")
ax.imshow(grid.T, origin="lower", cmap="inferno", norm=LogNorm(vmin=1e4, clip=True))
fig.savefig("load_halo_selection.png")
This produces the following image:
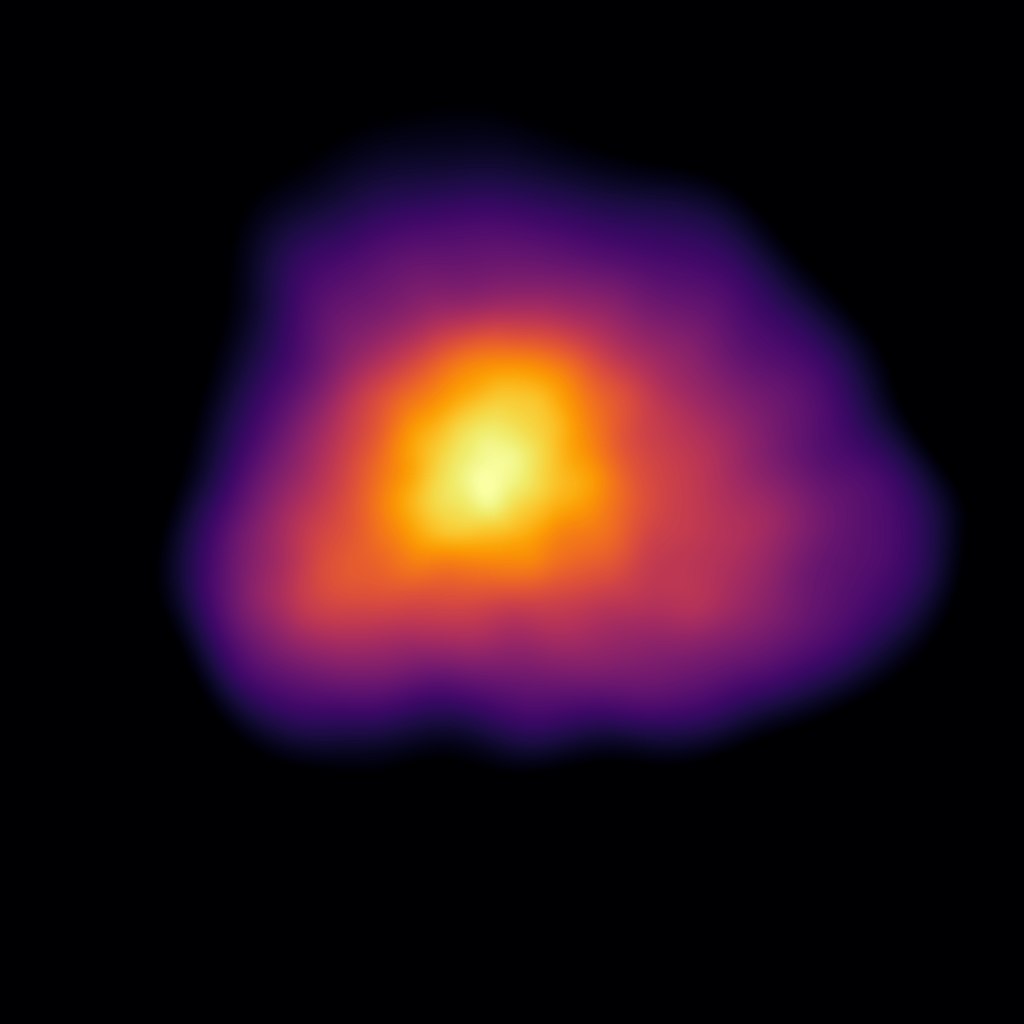
Then, finally, we can visualise only the bound particles, through the use of the mask
object that was returned when we initially extracted the swiftsimio dataset:
grid = project_gas_pixel_grid(data=data, resolution=1024, region=region, mask=mask.gas)
fig, ax = plt.subplots(figsize=(4, 4), dpi=1024 // 4)
fig.subplots_adjust(0, 0, 1, 1)
ax.axis("off")
ax.imshow(grid.T, origin="lower", cmap="inferno", norm=LogNorm(vmin=1e4, clip=True))
fig.savefig("load_halo_bound_selection.png")
Producing the following image:
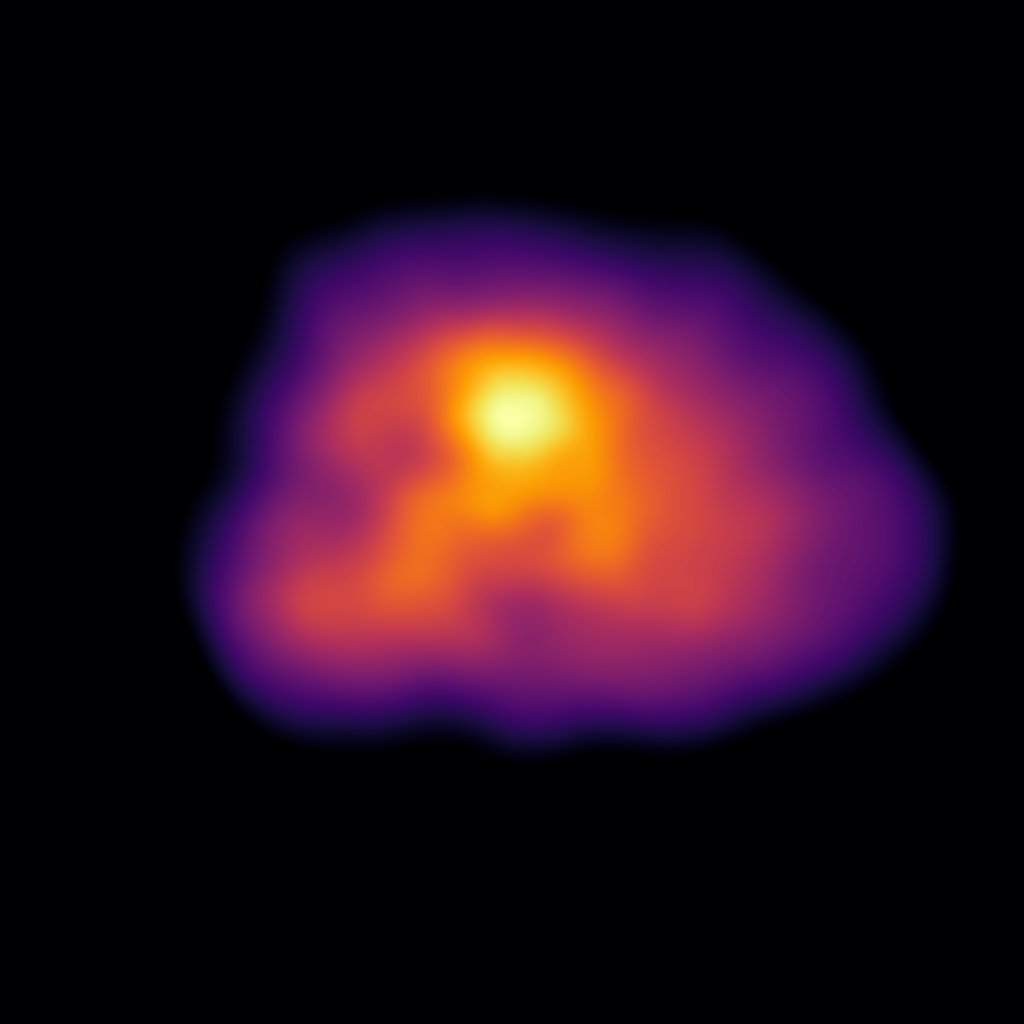
Hopefully, when you use this feature, you have more exciting data to use than the as-small-as-possible example that we show here!